| AlphaFold Solves Fundamental Biology Problem |
| Written by Mike James | |||
| Wednesday, 02 December 2020 | |||
|
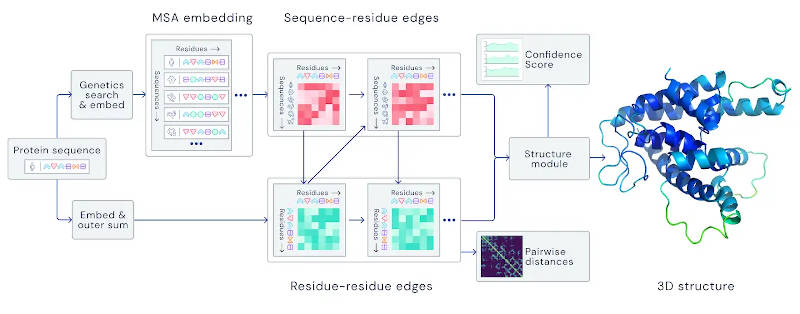
Back in 2018, we reported on DeepMind's attempts to create a neural network that would predict protein folding. Now we have the news that it so good that scientists are queuing up to use it. What is the problem, why is it important and what does it mean that a neural network can solve it where physics can't? One of the basic mechanisms of life is the way DNA not only codes for proteins, but actually makes them. The code in the DNA is expressed as a sequence of 20 amino acids. This chain of amino acids, the protein in question, is initally flat and unstructured, but what gives it many of its chemical and biological properties is the 3D shape it adopts. Each amino acid attracts other amino acids to different degrees. This means that when the chain is free to change its shape it wriggles around to find a lowest energy configuration. In a sense, the chain is like a spring that has been straightened and on release springs back to its natural lowest energy configuration. So far so good, but working out what the final shape should be is a very difficult combinatorial problem. You can attempt to solve it by doing classical chemical simulations trying to find the lowest energy configuration. The problem is that there are so many configurations it is difficult to find the one with lowest energy. For example, a chain of 100 units has has something in the region of 3198 configurations - a number that would take longer than the age of the universe to search - and this is a small protein. The following video gives you some idea of what protein folding is all about: The problem is clearly impossible and yet proteins, real proteins, solve the problem in a millisecond or less. Now Deep Mind's AlphaFold neural network has achieved a performance the puts it in the same class as direct determination of the structure using X-ray defraction, a process that can take years. The network was trained using a database of 170,000 proteins in a relatively short time on a set of TPUs: "16 TPUv3s (which is 128 TPUv3 cores or roughly equivalent to ~100-200 GPUs) run over a few weeks, a relatively modest amount of compute in the context of most large state-of-the-art models used in machine learning today." The structure of the neural network is also interesting. It attempts to learn a spatial graph where the nodes are the amino acids: "For the latest version of AlphaFold, used at CASP14, we created an attention-based neural network system, trained end-to-end, that attempts to interpret the structure of this graph, while reasoning over the implicit graph that it’s building. It uses evolutionarily related sequences, multiple sequence alignment (MSA), and a representation of amino acid residue pairs to refine this graph."
The important part of this description is "attention-based". This seems to be a general innovation in neural network design that makes the network capable of behavior more typical or recurrent networks - the attention substitutes for short-term memory. For the detail we will have to wait for the peer-reviewed paper that Deep Mind is currently preparing. The following video gives you some insight into what went on:
So how important is this? One answer is clear from a quote by Professor Andrei Lupas, Director of the Max Planck Institute for Developmental Biology: “AlphaFold’s astonishingly accurate models have allowed us to solve a protein structure we were stuck on for close to a decade, relaunching our effort to understand how signals are transmitted across cell membranes.” This really is a breakthrough and you can expect new techniques and new products as a result. The point is that we have the recipes for proteins in the form of DNA sequences, but until now we didn't know what shape an amino acid sequence would have. Now we know the recipe and can predict the end product. But wait there is a problem. Is this science? It clearly is technology, but having a neural network that predicts protein shape isn't the same as having a theory of how proteins fold. You can't even put "error bars" on how accurate any new prediction the network makes is. Can you trust it? For practical work. you can at least start out with the idea that it is correct and see if it makes sense. As far as theory goes, it really doesn't help - it's a back box that solves a problem without explaining how it does it. However, things are more subtle. If we go back to the idea that proteins fold in a millisecond there must be simpler organizing principles that mean that the protein doesn't try lots of possible foldings. It simply goes to the solution rather than searching a huge possible solution space, see Levinthal's paradox. Somewhere inside the structure of the network are the simpler rules that tell a typical protein how to fold without searching the space. This brings us to the important topic of neural networks that can explain their predictions. At the moment we have a very important technological advance that could well lead to new science. More InformationHigh Accuracy Protein Structure Prediction Using Deep Learning John Jumper, Richard Evans, Alexander Pritzel, Tim Green, Michael Figurnov, Kathryn Tunyasuvunakool, Olaf Ronneberger, Russ Bates, Augustin Žídek, Alex Bridgland, Clemens Meyer, Simon A A Kohl, Anna Potapenko, Andrew J Ballard, Andrew Cowie, Bernardino Romera-Paredes, Stanislav Nikolov, Rishub Jain, Jonas Adler, Trevor Back, Stig Petersen, David Reiman, Martin Steinegger, Michalina Pacholska, David Silver, Oriol Vinyals, Andrew W Senior, Koray Kavukcuoglu, Pushmeet Kohli, Demis Hassabis. Related ArticlesAlphaFold DeepMind's Protein Structure Breakthrough Fighting Coronavirus At Home With Exascale Power Nobel Prize For Computer Chemists To be informed about new articles on I Programmer, sign up for our weekly newsletter, subscribe to the RSS feed and follow us on Twitter, Facebook or Linkedin.
Comments
or email your comment to: comments@i-programmer.info |
|||
| Last Updated ( Wednesday, 02 December 2020 ) |